DeepMind, a subsidiary of Google, has launched an advanced AI system, AlphaProteo, designed to generate novel proteins with precise binding capabilities to target molecules. This development holds significant potential for advancing drug discovery and disease research.
AlphaProteo demonstrates exceptional performance in producing protein binders for various targets, including VEGF-A, a protein associated with cancer and complications related to diabetes. Notably, AlphaProteo is the first AI system to successfully design a protein binder for VEGF-A.
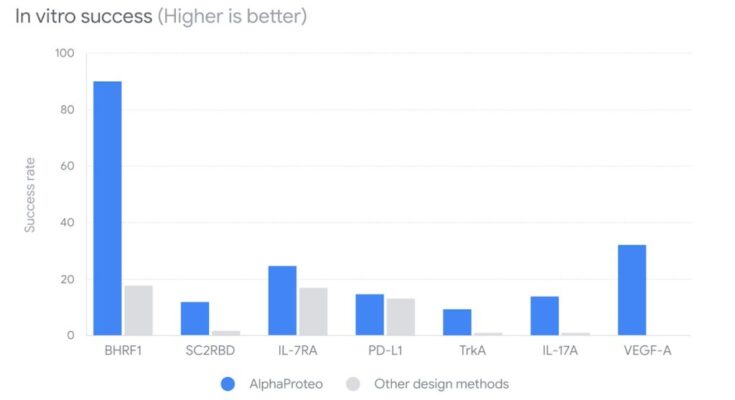
AlphaProteo stands out due to its remarkable experimental performance, where it surpassed existing methods by achieving significantly higher success rates and binding affinities up to 300 times stronger across seven different proteins. This marks a significant leap forward in the field of protein design, as binding affinity is a crucial factor in the effectiveness of protein-based therapies and molecular interventions.
Traditional protein engineering methods often rely on time-consuming and resource-intensive trial-and-error approaches. Scientists typically modify known proteins or use directed evolution techniques to optimize their binding properties, which can take months or even years. In contrast, AlphaProteo’s AI-driven approach accelerates this process by leveraging massive datasets and advanced machine learning algorithms to predict and design proteins with enhanced binding capabilities.
AlphaProteo was trained on extensive data from the Protein Data Bank, a comprehensive repository of experimentally determined protein structures, and over 100 million predicted structures generated by AlphaFold, DeepMind’s revolutionary AI tool for predicting protein folding. This vast dataset has enabled AlphaProteo to learn the intricate relationships between protein structures and their binding interactions, allowing it to accurately predict how proteins will bind to target molecules.
By analyzing the molecular structure of a target and identifying the most favorable binding sites, AlphaProteo designs custom proteins that can attach to those specific locations. This precision in design makes it a powerful tool for applications where strong and specific binding interactions are critical, such as in therapeutic drug development, where proteins need to bind to disease-causing molecules, or in diagnostics, where binding proteins are used to detect specific biomarkers.
Ultimately, AlphaProteo’s advanced training and design capabilities give it a distinct edge over traditional methods, promising to significantly reduce the time and cost associated with protein design, while improving the likelihood of successful outcomes in a variety of biomedical and research applications.
Researchers tested AlphaProteo by designing protein binders for various targets, including viral proteins and those related to cancer, inflammation, and autoimmune diseases. The results were highly promising, with AlphaProteo showing strong binding and high success rates.
For instance, 88% of its candidate proteins successfully bound to the viral protein BHRF1 in lab tests, and overall, its binders were 10 times stronger than current design methods.
AlphaProteo’s efficiency could greatly speed up protein binder experiments, advancing progress in various fields. However, it has limitations, such as its inability to design effective binders for TNFɑ, a protein linked to autoimmune diseases like rheumatoid arthritis.
To ensure responsible use, DeepMind is collaborating with experts and contributing to industry best practices, including working with the NTI’s AI Bio Forum.
As AlphaProteo continues to evolve, DeepMind is focused on collaborating with the broader scientific community to address some of the most urgent biological challenges. These partnerships will enable researchers to apply AlphaProteo’s capabilities to a wide range of critical areas, from identifying therapeutic targets to accelerating drug discovery and improving precision medicine. One of the key collaborators in this effort is Isomorphic Labs, a company dedicated to advancing drug design by leveraging AI-driven insights. Together, they aim to explore how AlphaProteo can streamline the development of new medications by designing proteins that specifically bind to disease-related molecules, offering new possibilities for treating conditions like cancer, infectious diseases, and autoimmune disorders.
However, despite the significant leap AlphaProteo represents in protein design, successful binding is only the first step in developing practical applications. Translating these findings into real-world therapies requires overcoming several bioengineering hurdles. For instance, after identifying a strong binder, researchers must ensure that the designed proteins are stable, non-toxic, and can function effectively in complex biological environments like the human body. This involves addressing issues such as protein folding, degradation, and immune response, which are critical for therapeutic success.
Additionally, while AlphaProteo has demonstrated impressive results in lab settings, scaling its application to broader drug development pipelines will require extensive testing, refinement, and regulatory approval. Each designed protein must undergo rigorous clinical trials to confirm its safety and efficacy in treating diseases. The complexity of human biology and the variability of disease mechanisms mean that even the most promising binders might face unforeseen challenges in clinical settings.
Furthermore, the integration of AlphaProteo into drug discovery workflows will likely require advancements in computational power and data integration. As AI-driven protein design tools like AlphaProteo generate vast amounts of data, there is a growing need for systems that can effectively manage, analyze, and interpret these insights in ways that inform clinical decision-making. This includes improving the ability to predict how designed proteins will behave in vivo and optimizing their interactions with other therapeutic agents.
In summary, while AlphaProteo represents a groundbreaking development in protein design, its true potential will be realized through continued research, collaboration, and the resolution of numerous bioengineering challenges. DeepMind’s partnerships with organizations like Isomorphic Labs and the scientific community at large will be critical in translating AlphaProteo’s innovations into practical, life-saving applications in medicine and beyond.
Nonetheless, DeepMind’s innovation presents substantial potential for accelerating progress in several critical domains, including drug development, disease research, and diagnostic technologies. Moreover, its applications extend beyond biomedicine, offering possibilities for advancements in agricultural science, such as the development of pest-resistant crops. The broad applicability of this technology underscores its capacity to address complex challenges across both healthcare and agricultural sectors, fostering interdisciplinary advancements and contributing to the development of more efficient, targeted solutions to pressing global issues.